Modeling Molecules with Recurrent Neural Networks
I enjoyed reading Andrej Karpathy’s The Unreasonable Effectiveness of Recurrent Neural Networks lately – it’s got some fascinating examples and some good explanations. I’ve been playing around with the char-rnn code from that post, and I want to share some of my experiments.

Chemical formulas and names
First experiment: I trained char-rnn on 3,892 real chemical compounds, both organic and inorganic, one formula and name per line, in randomized order.
C6H12O6, β-D-galactose
C4H8O2, ethyl acetate
C7H8N4O2, xantheose
C2H4O3, glycolic acid
CH5N, methylamine
C7H8N2S, phenylthiocarbamide
C17H20N4O6, riboflavin
C8H10, ortho-xylene
WOF2, tungsten(VI) oxytetrafluoride
PoO3, polonium trioxide
CuSe, copper(II) selenide
ErI3, erbium triiodide
NaHCO3, sodium bicarbonate
CH4, methane
...
After ten minutes of training, I had access to an excellent, excellent generator of completely fake chemistry jargon.
C4H8N2O2, thiochlorophecylene
AlCl6, aluminium cyandate bromide
C2H4BrO, bromo-3,5-cylohexanol
CH2CH3COOH, coltium carbide
C3I7N3, 2-buumenidine
C2H2O2, ethenol
RgClO4, malobium hexaxide
C2H5NO2, vinyl chloride
Na(VO3)2, sodium metatitanatedioxide
HOB2, deiopyre fluoride
C2H3CH3NO2, iomone acetic acid
CoCl2, cobalt(II) carbide
C19H14O, methoprepane
SnO3, strontium monoxide
C9H11NO, 1,3-pytan
Nr2, gannisil fluoride
C6H12, hadequine
C2HTi, chlorobenzelymethane
RCCl3, thyll
H2Br2, bromofluorosante
C15H17Cl2N2O3, lyctosin
C5H8ClN, anthalum-3-carboxyblue
N2N2O4, nitrogun grupiodide
C9H7N, γ-pinolylionine
C10C18O2, heptane
C32H21N4, gipsatedrale
SiBr4, strium tetralicolipethylachloradehydrame
C2H4ClO6, 2-chlorobromoprenidine
C7H4O4, methyl fluoride
FeS, vanady(III) fluoride
III2, irophiorite
...
Interestingly, the neural net is doing a good job of pairing organic-looking formulas with organic-looking names, and inorganic-looking formulas with inorganic-looking names. Other than that, it’s clearly not got enough data to have internalized the full periodic table of elements yet. I’m currently using this neural network to generate my Zephyr signatures.
Molecular structures
One thing that really struck me about the char-rnn neural network was its ability to remember its position in a stack. Karpathy gives numerous examples of training the neural network on different sources, then sampling the neural network to get randomly-generated output. Incredibly, when trained on Linux source code, on LaTeX, or on Markdown/XML from Wikipedia, the neural network creates syntactically-valid output. XML tags close. Parentheses match. This is quite beyond what Markov chains alone can do.
While building Carbonate, I spent a lot of time working with SMILES (Simplified Molecular-Input Line-Entry System), writing code to parse SMILES into molecules and code to convert molecules into SMILES. SMILES is a formalized scheme for converting molecules – which, by their nature, are nonlinear – into strings, a linear data structure. Here are some examples:
| CC(O)C, isopropanol | 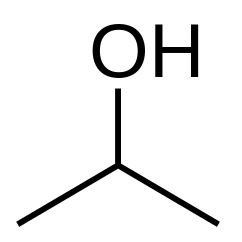 |
| C1CCCCC1, cyclohexane | |
| CC(=O)NCCC1=CNc2c1cc(OC)cc2, melatonin |  |
SMILES follows a formal grammar. It uses parentheses to branch off side-chains, as in CC(O)C for isopropanol. Rings are more complicated: to create the SMILES string for a ringed molecule, one must first cut some bonds so as to turn the molecule into a tree with no cycles; numerical indices are then used to link faraway atoms to each other in the resulting string. Chirality, cis/trans bonds, double or triple bonds, and aromaticity introduce yet further complications, each of which SMILES has ways of handling.
So:
- A valid SMILES string requires parentheses to match.
- char-rnn can create output with matching parentheses.
- ???
- Profit!
I acquired 1,200,000 different SMILES strings via ChemBank, and used them to train the neural net. Here is some of the output when I sample the result:
OCCCOc1ccc(cc1)C2=N[C@@](CCS(=O)(=O)c3ccccc3)([C@@H](O2)c4ccccc4)C(=O)NNCc5ccccc5C(F)(F)F COc1cc(cc(OC)c1OC)c2nnc(c3ccc(C)cc3)c4ccccc24 CN(C)CCCNC(=O)Oc1ccc2oc(C)c(C(=O)C)c2c1 OCCCCCCN1C(C(=O)N(CC=C)Cn2nnc3ccccc23)C4(CC[C@@H]5O4)[C@@H]([C@@H]5C(=O)N(CC=C)Cc6ccccc6)C1=O COc1ccc(CNNC(=O)[C@@]2(Cc3ccccc3)N=C(O[C@H]2c4ccc(Cl)cc4Cl)c5ccc(OCCCO)cc5)c1OC OCCCOc1ccc(cc1)C2=N[C@@](CCS(=O)(=O)c3ccccc3)([C@@H](O2)c4ccccc4)C(=O)NNCc5ccccc5C(F)(F)F CC(C)(C)OC(=O)CC[C@]1(N=C(O[C@@H]1c2ccccc2N=[N+]=[N-])c3ccc(OCCCO)cc3)C(=O)NCc4cccc(F)c4 CCOC(=O)[C@@H]1[C@H]2C(=O)N(CCCO)C(C(=O)N(CC=C)Cn3nnc4ccccc34)C52CC[C@@]1(CC)O5 COc1ccc(cc1O)[C@@H](O)c2ccc(OCCCCCC(=O)NC3CCCCC3)Cc2O OCCCOc1ccc(cc1)C2=N[C@@](Cc3ccccc3)([C@@H](O2)c4ccccc4)C(=O)NNCCc5cccc(c5)C(=O)NCCO CCCCCCCCCCCC(=O)N(CCc1ccccc1)C[C@]2(O)CC[C@H]3c4ccc(C[C@@H](O)CC/C(=C\CC[C@@]32C)/C)cc4C(=O)Cc5c(F)cccc5Cl COc1ccc(cc1O)[C@H](O)CC/C=C/C(=O)NO CC1(C)CCC[C@@H]2CCC1(C)N2C(=O)C3CCCCC3 CCOC(=O)[C@@H]1[C@H]2C(=O)N(CCCO)C(C(=O)N(CC=C)Cn3nnc4ccccc34)C52CC(C)[C@@]1(C)O5 CCCCCN1CC=C[C@@]23S[C@@H]4/C=C\CCCCOC(=O)[C@@H]4[C@H]3C(=O)N(CCCCCCO)C2C1=O Cc1cc(C)cc(OCCCCN2CCCCCC2)c1 CC[C@]1(CCC23O1)[C@H]([C@H]3C(=O)N(CCCCCO)C2C(=O)N(CC=C)Cn4nnc5ccccc45)C(=O)OCCCCC=C CC(C)OC(=O)[C@@H]1[C@@H]2CCC3(O2)C(N(CCO)C(=O)[C@H]13)C(=O)Nc4c(C)cccc4C COc1cc(ccc1O)C(=O)Nc2cccc(C)c2C COc1ccc(NC(=O)c2ccc(OC)cc2Br)cc1 CCCCCCCCCCCCCN/C=C/c1ccccc1 ...
Remarkably, these are all syntactically valid! The side-chains are sane, parentheses match, the ring-bonds match, and some of them even include chirality, cis/trans, and triple bonds! They're for fairly large molecules, but that's to be expected; my training data mostly consisted of large molecules. Because they're syntactically valid, they can be rendered into actual pictures using OpenBabel.
Next steps
Neural nets are fascinating. In addition to The Unreasonable Effectiveness of Recurrent Neural Networks, there are a few other posts that I read last summer – Google Research's Inceptionism, Felix Sun's DeepHear for composing and harmonizing music – that finally convinced me that neural networks are worth paying attention to. And char-rnn, in particular, has been simple to get working and pleasant to use.
If I decide to play around with this more, the next thing I'm going to do is to investigate whether a neural net could be trained to predict the outputs of some simple organic reactions. Modeling organic reactions was the bottleneck of the Carbonate project; the logic of each reaction had to be written and tested by hand. Using a neural net to learn the rules for reactions automatically might make for a better way forward.
